We are interested in resolving large data challenges in biology and medicine. With the use of deep learning we aim at developing new data-driven approaches to study of image and video data in biology. Our theoretical interests lie in using machine learning to find appropriate data representations for resolving specific scientific questions. We apply and develop computational and analytical solutions to questions in cancer and ageing research.
Located in the interdisciplinary environment of the CMMC, our group is involved in multiple close collaborations with partners from experimental and medical backgrounds. Strongly applied in biomedical research, we search for ways in which computational and in particular machine learning methods can advance resolving specific medical and biological questions. Currently we pursue two major projects.
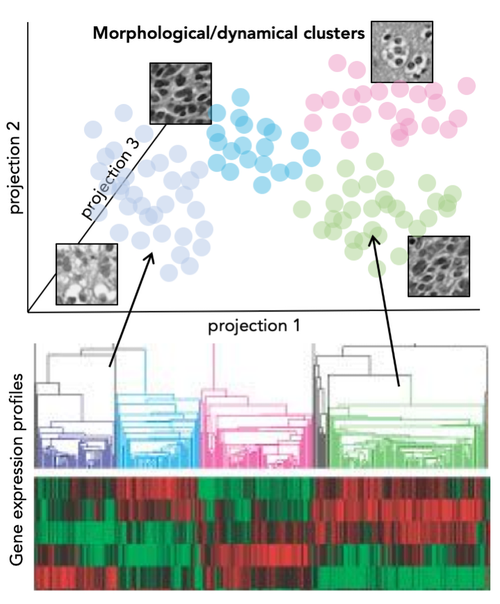
Highly differentiated diseases require large-volume, high-resolution datasets and novel analytical approaches to find recurring patterns in large spectrum of genetic and molecular variation. In collaboration with experimental groups at the Charité University Hospital Berlin we address the question of high molecular heterogeneity in Triple Negative Breast Cancer (TNBC). This most aggressive breast cancer subtype with high rates of recurrence and mortality offers currently no therapy options.
In this project we expand over the existing knowledge of the genetic heterogeneity between and within TNBC tumors, by investigating the differences in their transcriptome, proteome and morphology as well as the extracellular matrix components and immune cell environment. Our analysis is based on patient samples from prospective clinical trials at multiple levels. We strive to establish a novel type of biomarkers that instead of individual molecule levels will integrate multiple types of information and will be quantified with the use of machine learning and mathematical models.
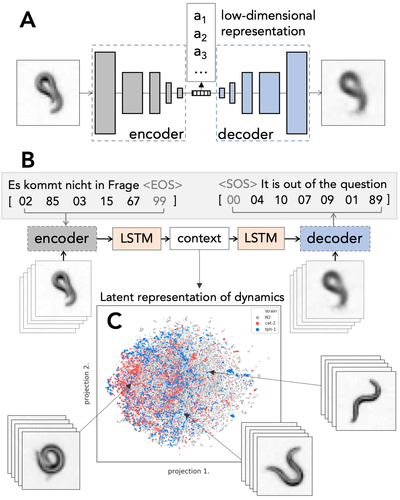
C. elegans, a tiny nematode worm, is used to study a broad range of questions in biology, from diseases to neural function. This apparently simple organism shows a broad repertoire of behaviors incomprehensible to human observer. These behaviors might be representative of its health and disease phenotype.
We employ deep learning methods to quantify and search for distinct motion patterns representative of worm molecular phenotype. The grand challenge of this project involves finding ways to represent worm posture and dynamics. Inspired by methods for language processing we aim to find analogous meaningful representations of words and sentences in the language of worm behavior.
The recent progress of both imaging and image analysis techniques are currently opening ways to elevate image data to a new scale in biological research. Whether in diagnostics of cancer or phenotyping of model organisms, we aim to establish methods and approaches allowing for broad, data scientific use of visual data in biology.
For further information please check the Bozek Laboratory for Data Sience of Images' webpage.

Data Analytics in Bioinformatics and
Center for Molecular Medicine Cologne