We are interested in the mechanisms that regulate chemical modifications of histone proteins. As signals that orchestrate gene expression, cell differentiation and many other processes, these reactions have to be faithfully controlled. Our biochemical understanding of the multiprotein complexes that catalyze these reactions is still very limited. Therefore, we aim to define aspects of their context dependent, local activity regulation using cryo-electron microscopy (cryo-EM), biochemical and cell biological approaches.
The many different cell types of our bodies share essentially the same genetic information, yet they differ dramatically in shape, behavior and function. A fundamental basis of these differences are the sets of genes that are transcribed (active) or repressed (inactive). Importantly, gene expression patterns, once set, can be passed on to daughter cells, thus allowing for stable cell identities within tissues and organs.
Transcriptional activity and many other DNA associated processes are controlled through epigenetic mechanisms. This term comprises regulatory processes that are not directly based on DNA sequence, but instead rely on interactions of proteins or nucleic acids with DNA, as well as their chemical modification.
Histone proteins form the core of nucleosomes, the basic packaging unit of DNA in the cell nucleus. Nucleosomes facilitate DNA compaction and organization, but are also essential hubs for transcriptional regulation.
The outcome of histone modifications depends on the kind of modification, the position of the amino acid that is being modified, and the context within the genome. Consequently, the enzymes that catalyze the attachment and removal of such modifications are important mediators of epigenetic processes. These chromatin modifying enzymes have to be present and active at the right time and place within the genome in order to perform their function.
In cancer, chromatin modifiers are frequently corrupted through mutations, chromosomal fusions or changes of their abundance, leading to aberrant gene expression patterns, loss of proliferation control or tissue invasive behavior.
In order to understand the function of chromatin modifiers in health and disease, we have to identify the molecular mechanisms that guide them to their targets within the genome and locally regulate their enzymatic activity. With such knowledge in hand, we hope to be able to develop novel, targeted strategies to manipulate chromatin modifiers for cancer therapy in the future.
Histone methyltransferases (HMTases) and demethylases are enzymes that attach and remove methyl groups from lysine or arginine residues, respectively. As all chromatin modifying enzymes, they are typically active as parts of multiprotein complexes. Subunits of these complexes and the enzymes themselves harbor interaction modules that help them recognize and respond to distinct features of their local chromatin environment. In our group, we employ chromatographic methods to isolate native chromatin modifying complexes from cultured human cells or heterologous expression systems such as insect, yeast and bacterial cultures. We are using single-particle cryo-electron microscopy (cryo-EM) and to visualize the three-dimensional structure of these molecular assemblies. The architecture of the full, native complexes will allow us to draw conclusions on the functional relationship between and within subunits.
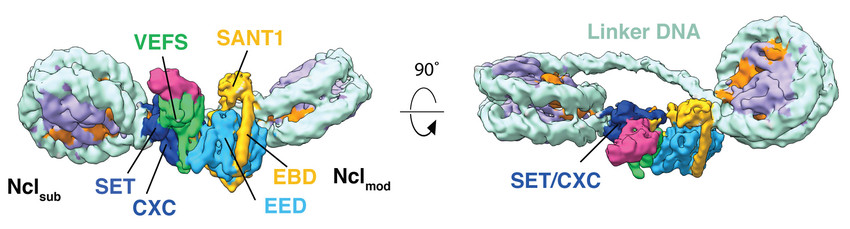
Figure 1: Cryo-EM reconstruction of human Polycomb Repressive Complex 2 (PRC2) in complex with a hetero-dinucleosome subtrate. The catalytic subunit EZH2 is allosterically activated by an H3K27me3 modified nucleosome (Nclmod) to methylate H3 of a neighboring nucleosome (Nclsub). Work performed in the laboratory of Eva Nogales (UC Berkeley) (Poepsel et al., 2018).
A central aspect of our studies are the interactions of HMTase and demethylase complexes with the chromatin environment, i.e. with nucleosomes and DNA. Using reconstituted nucleosomes as natural substrates and ligands will reveal molecular interactions that regulate and target their activity in the chromatin context. Structural studies will allow us to define these interactions in detail.
In vitro, the implications of our findings will be validated using mutational approaches coupled with biochemical activity and affinity assays. The functional significance is investigated by means of cellular systems of cell differentiation, such as mouse embryonic stem cells (mESCs) or human induced pluripotent stem cells (hiPSCs).
Together, these efforts aim at a better understanding of the enzymatic regulation of chromatin modifiers to control transciptional activation and repression, and cell proliferation.
By obtaining high resolution structural information on critical interactions defining the biology of chromatin modifying complexes, we aim to provide a basis for the rational design of drug candidates for the manipulation of their function in cancer and other diseases.
Further projects in our group focus on context dependent functions of epigenetic adaptor proteins, as well as other multi-subunit (nucleo-) protein assemblies implicated in disease processes.
For further information please check the Poepsel Laboratory For Structure and Biochemistry of Epigenetic Regulators' webpage.
Poepsel, S., Kasinath, V. and Nogales, E. (2018). Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nature Structural & Molecular Biology 25, 154-162.
Kasinath, V., Faini, M., Poepsel, S., Reif, D., Feng X.A., Stjepanovic, G., Aebersold, R. and Nogales, E. (2018). Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359, 940-944.
Kellogg, E.H., Hejab, N., Poepsel, S., Downing, K.H., DiMaio, F., Nogales, E. (2018). Near-atomic model of microtubule-tau interactions. Science 360(6394):1242-1246
Poepsel, S., Sprengel, A., Sacca, B., Kaschani, F., Kaiser, M., Gatsogiannis, C., Raunser, S., Clausen, T., Ehrmann, M. (2015). Determinants of amyloid fibril degradation by the PDZ protease HTRA1. Nature Chemial Biology 11, 862-9.
Kasinath, V*, Poepsel, S.*, and Nogales, E (2019). Recent Structural Insights into Polycomb Repressive Complex 2 Regulation and Substrate Binding. (REVIEW) Biochemistry 58(5):346-354. * equal contributions

Center for Molecular Medicine Cologne | Lab. of Structure and Biochemistry of Epigenetic Regulators | CMMC Research Building
CMMC - PI - JRG 11
former CMMC - Co-PI - A 12
+49 221 47896987
Center for Molecular Medicine Cologne | Lab. of Structure and Biochemistry of Epigenetic Regulators | CMMC Research Building
Robert-Koch-Str. 21
50931 Cologne