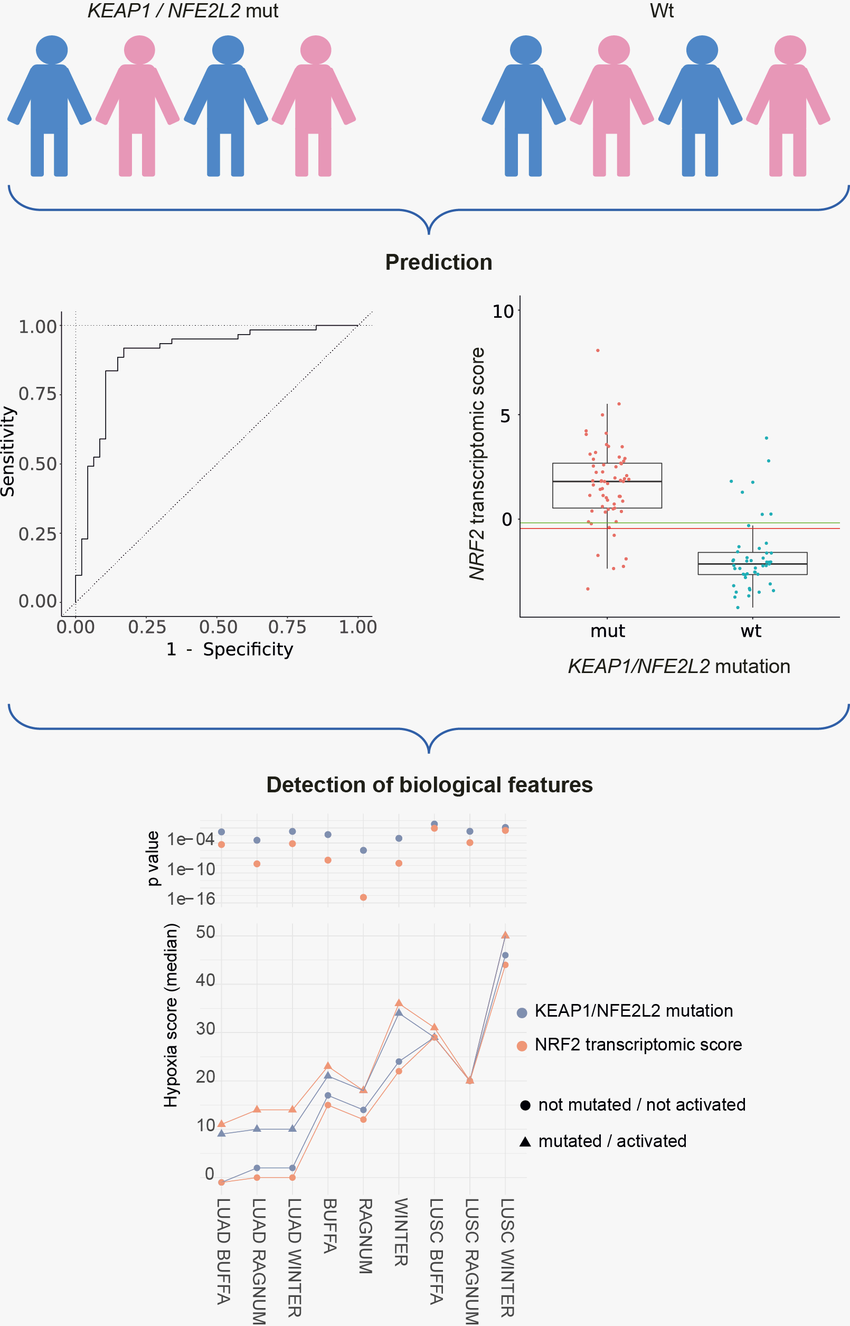
Lung cancer is the leading cause of cancer-related death with lung adenocarcinoma (LUAD) as the most frequent subtype. KEAP1 is mutated in 15-20% of LUAD, a patient group with particularly poor prognosis. In situations of high reactive oxygen species (ROS), NRF2 initiates the expression of pro-survival genes that quench ROS. In KEAP1 mutated LUAD, the NRF2 (gene name: NFE2L2) pathway is constitutively active, allowing the cancer to cope with high ROS levels. This adaptation makes cancer cells resistant to radio- and chemotherapy. There is an urgent clinical need to develop new treatment strategies for patients with KEAP1 mutated LUAD. A promising strategy is to inhibit the NRF2 pathway. We evaluate the pathogenic role of individual KEAP1 mutations by screening our clinical cohort using immunohistochemistry and an expression assay that identifies tumors with NRF2 pathway activity. In addition, we will test for the functional consequences of specific KEAP1 mutations in vitro. Further, we will search for genomic alterations other than KEAP1/NFE2L2 mutations that can activate the NRF2 pathway. Finally, we will establish model LUAD cell lines with inducible KEAP1 activity to screen drugs with effects specific for KEAP1 mutated tumors. The effect of candidate drugs will be further characterized by proteomics and metabolomics. Overall, we will pave the way to new treatment strategies for a large group of LUAD patients with poor prognosis.
We will apply this concept to treatment naïve EAC biopsies and normal tissues and will systematically identify cell-type specific alterations that correspond to treatment response and relapse. We will validate our findings using a large collection of EAC tissues for targeted assays and by experiments in tissue culture. Overall, we will be able to identify functional changes in CAFs and other tumor components that will help to explain the clinical course of an EAC patient and that provide new opportunities for intervention.
KEAP1 mutated LUAD has a poor prognosis. We will establish a better definition of NRF2 activated tumors than relying on the non-silent mutations in KEAP1 or NFE2L2 and will identify drugs that target the NRF2 pathway. Such drugs bear a great potential to improve treatment response in the large group of advanced stage non-small cell lung cancer patients with NRF2 pathway activating mutations, a patient subgroup with particularly high medical need for better therapies (in Germany: 2,500 per year).

Institute for General Pathology and Pathological Anatomy
CMMC - PI - A 06
show more…
+49 221 478 85643
Institute for General Pathology and Pathological Anatomy
Joseph-Stelzmann-Str. 26
50931 Cologne
https://pathologie.uk-koeln.de/forschung/genomische-pathologie-ag-hillmer/