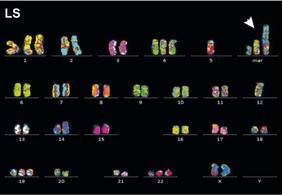
Multicolor FISH analysis of seismic amplification presenting as giant marker chromosomes or neochromosomes (NC) in metaphase spreads of cell line LS. Arrowhead indicates the NC. Scale bars, 10 μm. CEP2, centromere protein of chromosome 2; XCP12, whole chromosome 12 paint (see Figure 2).

Dr. Carolina Rosswog and Prof. Dr. Matthias Fischer Our therefore provides novel insights into the mechanisms of how human tumors acquire genomic amplifications as major drivers of oncogenic transformation. An important finding for the development of preventive measures and new treatment strategies. (Foto: Michael Wodak)
Cancer is caused by changes in the genome of body cells. In many cases, these are selective gene changes, but there is often also a multiplication ("amplification") of cancer genes. These genes are then not present in two copies, as in normal cells, but in 20, 100 or even 500 copies, which in turn leads to a strong increase in their activity and thus contributes to the development of tumors. The mechanisms behind the evolution of complex genomic amplifications in cancer have been poorly understood to date.
Scientists from the Dept. of Experimental Pediatric Oncology, the Dept. of Translational Genomics, the Center for Molecular Medicine Cologne, Faculty of Medicine, University of Cologne and the University Hospital Cologne and among other collaborative partners investigated the development of highly complex amplifications in cancer cell. In a cooperative approach the scientists identified a new type of amplification, termed as “seismic amplification”, which is characterized by multiple rearrangements and numerous genomic segments amplified at distinct levels. The phenomenon of seismic amplification occurs frequently in cancer cells. The results of the cooperative study have now been published in the renowned scientific journal Nature Genetics - Chromothripsis followed by circular recombination drives oncogene amplification in human cancer (Nat Genet. 2021 Nov 15) - https://doi.org/10.1038/s41588-021-00951-7.
In addition to extreme copy number fluctuations, the researchers found in these amplifications a highly altered sequence of the normal DNA sequence with numerous rearrangements within the DNA strand. Because the chaotic picture of these amplifications resembles the waves of a seismogram during an earthquake, the researchers have given these changes the name "seismic amplifications." By analyzing the structure of these amplifications in detail, they were able to decipher a mechanism that shows how these changes occur in tumor evolution.
Initially, part of one or more chromosomes breaks down into many small fragments, an event called "chromothripsis" that is known to occur in the genome of malignant tumors. Some of the resulting DNA fragments subsequently join together to form DNA rings, while other parts are lost. Within the DNA rings, a so-called circular recombination subsequently takes place, resulting in numerous rearrangements and an irregular duplication of DNA fragments, giving rise to the complex and characteristic picture of seismic amplifications. The degree of complexity depends on how many rounds of circular recombination occur. The researchers were also able to show that sometimes the DNA rings re-integrate into chromosomes and stabilize in this way. In other cases, the DNA rings remain in their circular form, are further amplified as such, and eventually hundreds of them are present in the nucleus of tumor cells.
"We first investigated neuroblastomas," reports Dr. Carolina Rosswog, first author of the study and clinician-scientist at the Dept. of Experimental Pediatric Oncology, the Else Kröner Forschungskolleg Cologne and CMMC. "These are malignant tumors of the peripheral nervous system that occur in early childhood. Seismic amplifications could be detected in about a quarter of the neuroblastomas and often involved known cancer genes, such as MYCN, MDM2 and CDK4. On this basis, the researchers examined nearly 2,700 tumors of other types of cancer and found that about 10 percent of all cases showed "earthquakes" in the genome. As a result of these changes - similar to neuroblastoma - there is usually a strong activation of cancer. Detailed reconstruction of the development of seismic amplification revealed a stepwise evolution behind these alterations, starting with a singular event of chromothripsis, followed by formation of extrachromosomal circular DNAs that subsequently underwent repetitive rounds of circular recombination", Rossweg comments further.
Due to the high complexity of seismic amplifications, extensive and computationally intensive simulations were performed. Detailed reconstruction of the development of seismic amplification revealed a stepwise evolution behind these alterations, starting with a singular event of chromothripsis, followed by formation of extrachromosomal circular DNAs that subsequently underwent repetitive rounds of circular recombination. In overt tumors, the resulting amplified regions persisted as extrachromosomal DNA circles or had reintegrated into the genome, and did not substantially change at relapse, suggesting that seismic amplifications are stable events.
“We found that seismic amplifications were associated with the occurrence of chromothripsis across cancer types and chromosomes, thus supporting our hypothesis of chromothripsis as the initiating event in the evolution of these alterations”, says Dr. Christoph Bartenhagen, computer scientist and co-first-author of the study. "We also observed that genomic loci affected by seismic amplification frequently covered proto-oncogenes, such as CCND1 (chromosome 11q), CDK4, MDM2 (both chromosome 12q), and ERBB2 (chromosome 17q). Importantly, seismic amplification led to excess copy numbers and massive over-expression of these genes in comparison to other amplification types or non-amplified states", Bartenhagen adds.
The data provide strong evidence that seismic amplification is a frequent event in cancer and follows a common evolutionary path, which is based on a stepwise evolution involving an initial chromothripsis event and subsequent recombination of extrachromosomal circular DNAs.
“Importantly, our data corroborate that seismic amplification defines a subgroup of highly complex genomic alterations that is not covered by current categories of structural variations in cancer”, explains Prof. Dr. Matthias Fischer (last-author of the study), head of the Dept. of Experimental Pediatric Oncology and a CMMC research group. The study therefore provides novel insights into the mechanisms of how human tumors acquire genomic amplifications as major drivers of oncogenic transformation - an important finding for the development of preventive measures and new treatment strategies."
Scientific Contacts:
Prof. Dr. Matthias Fischer
Dept. of Experimental Pediatric Oncology
University of Cologne, Faculty of Medicine and University Hospital Cologne
matthias.fischer[at]uk-koeln.de
Prof. Dr. Martin Peifer
Dept.of Translational Genomics
University of Cologne, Faculty of Medicine and University Hospital Cologne
mpeifer@uni-koeln.de
Original paper:
Chromothripsis followed by circular recombination drives oncogene amplification in human cancer
Carolina Rosswog1,2,3,15, Christoph Bartenhagen1,2,15, Anne Welte 1,2, Yvonne Kahlert1,2, Nadine Hemstedt 1, Witali Lorenz1, Maria Cartolano 2, Sandra Ackermann 1,2, Sven Perner 4,5, Wenzel Vogel 4,5, Janine Altmüller 6,7,8, Peter Nürnberg2,6, Falk Hertwig1, Gudrun Göhring9, Esther Lilienweiss10, Adrian M. Stütz11, Jan O. Korbel 11, Roman K. Thomas12,13,14, Martin Peifer 2,12 and Matthias Fischer 1,2
Nat Genet. 2021 Nov 15. https://doi.org/10.1038/s41588-021-00951-7. Online ahead of print. PMID: 34782764
https://www.nature.com/articles/s41588-021-00951-7
Authors in bold contributed equally.
Institutions of the authors:
1 - Dept. of Experimental Pediatric Oncology, University Children’s Hospital of Cologne, Faculty of Medicine and Univ. Hospital Cologne, Cologne,
DE
2 - Center for Molecular Medicine Cologne (CMMC), University of Cologne, Faculty of Medicine and Univ. Hospital Cologne, Cologne, DE
3 - Else Kröner Forschungskolleg Clonal Evolution in Cancer, University Hospital Cologne, Cologne, DE
4 - Institute of Pathology, University of Luebeckand University Hospital Schleswig-Holstein, Campus Luebeck, Luebeck, DE
5 - Pathology Research Center Borstel, Leibniz Lung Center, Borstel, DE
6 - Cologne Center for Genomics (CCG), University of Cologne, Faculty of Medicine and University Hospital Cologne, Cologne, DE
7 - Berlin Institute of Health at Charité, Berlin Germany, Core Facility Genomics, Berlin, DE
8 - Max Delbrück Center for Molecular Medicine, Berlin, DE
9 - Dept. of Human Genetics, Hannover Medical School, Hannover, DE
10 - Dept. of Internal Medicine, Univ. of Cologne, DE
11 - European Lab. Genome Biology Unit, Heidelberg, Germany
12 - Dept.of Translational Genomics, Medical Faculty, University of Cologne, Cologne, DE
13 - Dept. of Pathology, University of Cologne, Cologne, DE
14 - German Cancer Consortium (DKTK), German Cancer Research Center (DKFZ), Heidelberg, DE
Press Reselase from the University Hospital Cologne:https://www.uk-koeln.de/uniklinik-koeln/presse-mediathek/presse/details/erdbeben-im-erbgut-von-krebserkrankungen/