T cells dynamically expand and contract following antigen-specific stimuli and microenvironmental cues. As a result, immunological memory protects us from following antigen-specific attacks. Alterations of the mechanisms regulating this process are responsible for autoimmunity or impaired immune protection. Mitochondrial fitness centrally regulates memory T cell differentiation and function. It provides metabolic reserve capacity to allow long lasting survival of memory T cells. It also controls critical steps in epigenetic remodeling, chromatin accessibility and transcriptional profile of T cells by producing and buffering Acetyl-CoA and other metabolites. By combining the analysis of gene expression and chromatin accessibility of effector and memory T cells, we identified previously uncharacterized common hits regulated during memory T cell differentiation. Of note, these hits share a signature for some specific transcription factors, whose role in the immune system has never been studied. We hypothesize that these transcription factors might integrate metabolic and epigenetic signals to establish persistent immunological memory. By complementing metabolic and immunologic phenotyping with cutting edge sequencing approaches, the project will reveal novel insights into the epigenetic and metabolic network necessary to establish long-term immune memory and surveillance against cancer and recurrent infections.
Establishing long-term immune memory able to recognize and fight previously encountered antigens is central to the success of any vaccination campaigns or cancer immunotherapy. Nevertheless, the mechanisms underlying induction and maintenance of immunological memory still remain elusive. Our line of research aims at identifying and characterize metabolic and epigenetic checkpoints to be exploited in cancer immunotherapy or to improve current and future vaccine regimens.
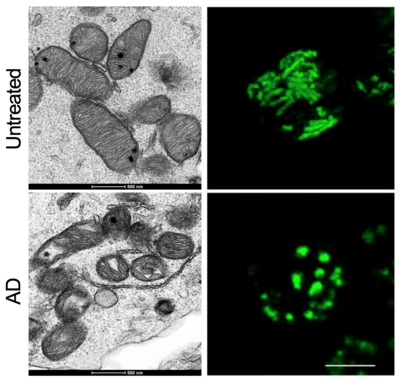
In this regard, metabolism has a central role in memory T cell generation. It is therefore not surprising that patients affected by mitochondrial diseases (MDs) also show immune defects linking systemic metabolism to immune function alteration (Corrado and Pearce, 2022). Up to half of the patients with MDs experience recurrent or severe upper respiratory tract infections, often resulting in life-threatening conditions. This percentage increases to almost 90% of pediatric MD patients. We hypothesize and investigate how mitochondrial and metabolic deficiencies might underlie and explain (at least partially) the high variability in terms of prognosis after an infection or after the diagnosis of an autoimmune disorder.

CECAD Research Center
CMMC - PI - B 03
CMMC - PI - CAP 21
CMMC - Co-PI - B 02
+49 221 478 84167
CECAD Research Center
Joseph-Stelzmann-Str. 26
50931 Cologne
PhD students:
Regina Annamaria
Sara Gjurgji
Master students:
Irma Alibashikj
Sivanesan Pujyanathan
Keanu Haenen
Yevheniia Minchuk
Technician:
Jessica Büchel