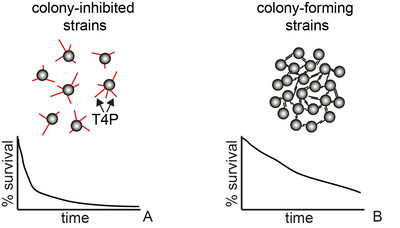
Aggregation into colonies and biofilms can protect bacteria from antibiotic treatment. Treatment of biofilm-related infections requires a detailed understanding of the molecular mechanisms of antibiotic tolerance. This project aims at understanding how colony formation protects the human pathogen Neisseria gonorrhoeae from antibiotic treatment. We will focus on the interplay between the physical properties of gonococcal colonies and antibiotic susceptibility.

According to WHO, gonorrhea is currently the second most common STI worldwide. Vaccination is not available and the probability of failure of treatment with antibiotics is rising rapidly (1). Due to its rapid acquisition of antibiotic resistance, (multi-) drug resistant N. gonorrhoeae have been termed an urgent threat by the US Center for Disease Control. Some aspects of drug resistance are understood at the genetic level, but the effect of biofilm formation and its properties on efficiency of treatment remains poorly understood. This project aims at elucidating potential mechanisms of antibiotic tolerance in gonococcal biofilms.

Institute for Biological Physics - Center for Molecular Biosciences
CMMC - PI - B 08
berenike.maier[at]uni-koeln.de
show more…+49 221 470 8046
Institute for Biological Physics - Center for Molecular Biosciences
Zülpcher Str. 47a
50674 Cologne
PostDoc
Dr. Marc Hennes
Dr. Stephan Wimmi
Dr. Melih Yüksel
PhD student
Mona Förster
Sebastian Kraus-Römer
Ariana Leu
Isabel Rathmann
Aurelio Scarpone
Isabelle Wielert
Master students
Mahak Sadwani
Paul Schiefer
Technician
Thorsten Volkmann

Institute for Medical Microbiology, Immunology and Hygiene
CMMC - Co-PI - B 08
show more…+49 221 478 32011
Institute for Medical Microbiology, Immunology and Hygiene
Goldenfelsstraße 19-21
50935 Cologne
PostDoc
Dr. Kyriaki Xanthopoulou
PhD student
Dr (med) Annika Y. Claßen
Dr-med students
David Bogdanski
Aime Picornell
Technician
Rocío Arazo del Pino
Tessa Burgwinkel
Vivien Persy