Nonsense-mediated mRNA decay (NMD) is a crucial quality control mechanism in eukaryotic cells that safeguards the fidelity of protein synthesis. NMD selectively eliminates messenger RNAs (mRNAs) that carry premature translation termination codons, commonly referred to as NMD substrates. Thereby NMD prevents the production of truncated polypeptides that may have toxic effects on cellular functions. The process of NMD is executed by a complex machinery composed of RNA-binding proteins and NMD factors that work together to recognize, and degrade NMD substrates.
Mutations in genes involved in the NMD mechanism have been linked to neurodevelopmental disorders (NDDs). For example, pathogenic variants in the gene encoding the NMD activator UPF3B have been identified as a cause of X-linked intellectual disability (ID). Similarly, deletions of genomic regions containing the gene for the NMD factor UPF2 have been associated with ID and other NDDs. Furthermore, point mutations in the NMD regulators SMG8 and SMG9 have been linked to cardiac and cerebral malformations. Despite these observations, the precise mechanisms by which NMD mutations contribute to NDDs remain unclear.
Here, we propose to dissect the function of NMD during brain development. We will use hiPSCs to knock out UPF3B by genome editing and/or alter the expression of the paralog UPF3A, which modulates the disease phenotype in patients with UPF3B mutations. In addition, we will up- and downregulate the global NMD efficiency in hiPSCs by different genetic interventions and pharmacological treatments.
As one main approach we will use the generation of brain organoids, which provide an excellent model system to investigate the mechanisms of brain development, including the role of NMD. We will investigate the effects of different NMD manipulations on the formation, composition, and complexity of brain organoids.
Our project aims to dissect the function of NMD during brain development and shed light on the mechanisms by which mutations in NMD genes contribute to NDDs. By studying the molecular and cellular effects of NMD inhibition and activation, we hope to identify potential therapeutic targets for NDDs that are linked to NMD dysfunction.
Between 1 and 5% of children worldwide are diagnosed with various neurodevelopmental disorders (NDDs). Genetic studies suggest a high heritability, but the clinical symptoms of NDDs are often heterogeneous. To diagnose and accelerate the treatment of genetic NDDs, it is essential to define the full spectrum of defective molecular pathways. Therefore, our project aims to study the role of nonsense-mediated decay (NMD) in brain development to dissect the molecular basis of NMD-dependent NDDs.
Linder B, Fischer U, Gehring NH (2015) mRNA metabolism and neuronal disease. FEBS Lett 589: 1598-1606
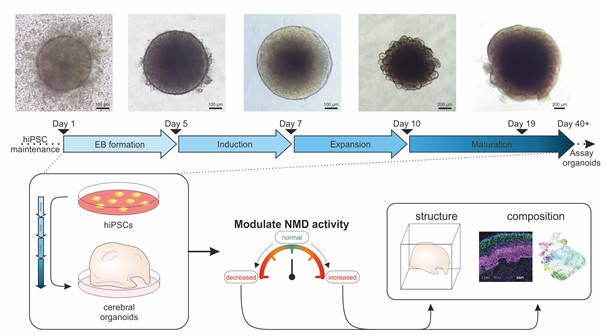
Fig.2: Modulating NMD activity in brain organoids and analyzing the effects.
Top: Representative images showing the formation, expansion, and maturation of brain organoids generated in the Gehring lab.
Bottom: Brain organoids will be generated from hiPSCs using the methodology described above. Different treatments and genetic interventions will be applied to modulate global NMD activity, as depicted schematically. The structure and composition of the organoids will be compared using transcriptomic and proteomic analyses, imaging techniques, and functional assays.

Institute for Genetics - Center of Molecular Biosciences
CMMC - PI - C 05
show more…+49 221 470 3873
Institute for Genetics - Center of Molecular Biosciences
Zülpcher Str. 47a
50674 Cologne
PostDoc:
Dr. Volker Böhm
PhD students:
Damaris Wallmeroth
Sabrina Kückelmann
André Müller
Master student:
Sophie Theunissen
Technician
Silke Modersohn
Juliane Hancke
Vanessa Paszella