Our lab is interested in understanding the basic molecular rules by which a cell defines and maintains its identity and function. Our main focus is on understanding the molecular basis of programming and reprogramming of cell-fate decisions during embryogenesis, homeostasis and aging. Additionally, we focus on devising molecular strategies to ‘hack’ these genetic networks that programs cell-fates to induce regenerative responses upon injury.
The human body is composed of about hundreds of different cell types. The identity and function of these distinct cell types are precisely programmed by the regulatory networks encoded in the 3 billion base pairs of DNA that constitute the human genome. While 60% of our genome is transcribed, less than 2% of it is translated to proteins. In contrast to previous assumptions, this suggests that a significant majority of the regulatory information from the genome functions as RNAs, termed non-coding RNAs. Emerging evidences suggest that a substantial portion of these non-coding transcripts control myriad biological processes ranging from development to disease, establishing the vital role played by these RNA regulatory elements.
A temporal decline in the functional and molecular integrity of an organism defines its ageing process. While multiple hallmarks of ageing have recently been enumerated, our understanding of their functional, molecular and temporal hierarchy remains incomplete.
The significance of these hallmarks in the ageing process vary in a tissue/ cell type specific manner. Considering the physiological differences between organs, it is conceivable that their susceptibility to these distinct triggers of ageing can differ.
Therefore, it is important to delineate those mechanisms that influence the ageing process of specific organs to better understand organismal ageing. We investigate evolutionarily conserved mechanisms that leads to aging of the heart.
Heart failure is a leading cause of mortality and morbidity in the developed world, partly because of the minimal regenerative ability of the mammalian heart. However, several fish and amphibians do possess dramatic ability to regenerate damaged organs, including heart. We use a combinatorial approach including regeneration competent models, state-of-the art stem cell-based models and systems biology approaches to understand the hidden regulatory layers enabling organ/ tissue regeneration.
Precision in gene expression is essential to establish and maintain a cellular identity in embryonic development. Therefore, key regulatory mecha-nisms are in place at every step of develop-mental cell-fate transitions, ranging from transcriptional control to proteostasis. Interestingly, recent studies suggest that up to 60% of gene expression is regulated at the level of translation.
Highlighting its importance in developmental gene regulation, slight alterations in mechanisms regulating translation leads to severe developmental defects or embryonic lethality. However, the role of translational regulation in shaping developmental gene expression landscape is poorly understood.
Additionally, transient alterations in the translation of specific mRNAs can have profound changes in molecular homeostasis. Using human pluripotent stem cell based models we investigate currently unknown mediators that regulate translation to program cell-fate.
Cardiac failure is a burgeoning public health issue predicted to reach epidemic proportion as the current population ages. Compounding reason for this is the inability of the adult mammalian heart to elicit effective regeneration. In our lab we intent to comprehend the molecular basis of cardiac development and regeneration, widening the array of therapeutic opportunities.
2. Ahuja G, Bartsch D, Yao W, Geissen S, Frank S, Aguirre A, Russ N, Messling J, Dodzian J, Lehmann K, Vargas N, Muck JS, Brodesser S, Baldus S, Sachinidis A, Hescheler J, Dieterich C, Trifunovic A, Papantonis A, Petrascheck M, Klinke A, Jain M, Valenzano D, Kurian L. Loss of genomic integrity by lysosphingolipid imbalance drives ageing in the heart. EMBO reports2019 Mar 18.
3. Chao-Chung Kuo, Sonja Hanzelmann, Nevcin Sentürk, Stefan Frank, Barna Zajzon, Jens-Peter Derks, Vijay Suresh Akhade, Gaurav Ahuja, Chandrasekhar Kanduri, Ingrid Grummt, Leo Kurian, and Ivan G. Costa. Detection of RNA–DNA binding sites in long noncoding RNAs. Nucleic Acids Research.2019 Jan 30.
4. Bender D, Kaczmarek AT, Santamaria-Araujoa JA, Stuevec B, Waltzc S, Bartsch D, Kurian L, Cirak S, Schwarz G. Impaired mitochondrial maturation of sulfite oxidase in a patient with severe sulfite oxidase deficiency. Human Molecular Genetics 2019 May 25.
5. Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, Hertwig F, Wernig-Zorc S, Miranda C, Deland L, Volland R, Bartenhagen C, Bartsch D, Bandaru S, Engesser A, Subhash S, Martinsson T, Carén H, Akyürek LM, Kurian L, Kanduri M, Huarte M, Kogner P, Fischer M, Kanduri C. Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 Determines Neuroblastoma Susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell.2018 Mar 12;33(3):417-434.e7
6. Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Nelles DA, Rodriguez Esteban C, Campistol JM, Yeo GW, Izpisua Belmonte JC Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 2015;7;131(14):1278-90.
Ahuja, G., Bartsch, D., Yao, W., Geissen, S., Frank, S., Aguirre, A., Russ, N., Messling, J.E., Dodzian, J., Lagerborg, K.A., Vargas, N.E., Muck, J.S., Brodesser, S., Baldus, S., Sachinidis, A., Hescheler, J., Dieterich, C., Trifunovic, A., Papantonis, A., Petrascheck, M., Klinke, A., Jain, M., Valenzano, D.R., and Kurian, L. (2019). Loss of genomic integrity induced by lysosphingolipid imbalance drives ageing in the heart. EMBO Rep 20.
Bender, D., Kaczmarek, A.T., Santamaria-Araujo, J.A., Stueve, B., Waltz, S., Bartsch, D., Kurian, L., Cirak, S., and Schwarz, G. (2019). Impaired mitochondrial maturation of sulfite oxidase in a patient with severe sulfite oxidase deficiency. Hum Mol Genet10.1093/hmg/ddz109.
Frank, S., Ahuja, G., Bartsch, D., Russ, N., Yao, W., Kuo, J.C., Derks, J.P., Akhade, V.S., Kargapolova, Y., Georgomanolis, T., Messling, J.E., Gramm, M., Brant, L., Rehimi, R., Vargas, N.E., Kuroczik, A., Yang, T.P., Sahito, R.G.A., Franzen, J., Hescheler, J., Sachinidis, A., Peifer, M., Rada-Iglesias, A., Kanduri, M., Costa, I.G., Kanduri, C., Papantonis, A., and Kurian, L. (2019). yylncT Defines a Class of Divergently Transcribed lncRNAs and Safeguards the T-mediated Mesodermal Commitment of Human PSCs. Cell Stem Cell 24, 318-27 e8.
Kuo, C.C., Hanzelmann, S., Senturk Cetin, N., Frank, S., Zajzon, B., Derks, J.P., Akhade, V.S., Ahuja, G., Kanduri, C., Grummt, I., Kurian, L., and Costa, I.G. (2019). Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res 47, e32.
Frank S, Ahuja G, Bartsch D, Russ N, Yao W, Kuo JC, Derks JP, Akhade VS, Kargapolova Y, Georgomanolis T, Messling JE, Gramm M, Brant L, Rehimi R, Vargas NE, Kuroczik A, Yang TP, Sahito RGA, Franzen J, Hescheler J, Sachinidis A, Peifer M, Rada-Iglesias A, Kanduri M, Costa IG, Kanduri C, Papantonis A, and Kurian L (2018). yylncT Defines a Class of Divergently Transcribed lncRNAs and Safeguards the T-mediated Mesodermal Commitment of Human PSCs. Cell Stem Cell10.1016/j.stem.2018.11.005.
Keller T, Wengenroth L, Smorra D, Probst K, Kurian L, Kribs A, and Brachvogel B (2018). Novel DRAQ5/SYTOX(R) Blue Based Flow Cytometric Strategy to Identify and Characterize Stem Cells in Human Breast Milk. Cytometry B Clin Cytom10.1002/cyto.b.21748.
Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, Hertwig F, Wernig-Zorc S, Miranda C, Deland L, Volland R, Bartenhagen C, Bartsch D, Bandaru S, Engesser A, Subhash S, Martinsson T, Caren H, Akyurek LM, Kurian L, Kanduri M, Huarte M, Kogner P, Fischer M, and Kanduri C (2018). Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 Determines Neuroblastoma Susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell 33, 417-434 e417.
Palanimurugan R, Godderz D, Kurian L, and Jurgen Dohmen R (2018). Analysis of Cotranslational Polyamine Sensing During Decoding of ODC Antizyme mRNA. Methods Mol Biol 1694, 309-323.
Ju Lee H, Bartsch D, Xiao C, Guerrero S, Ahuja G, Schindler C, Moresco JJ, Yates JR, 3rd, Gebauer F, Bazzi H, Dieterich C, Kurian L, and Vilchez D (2017). A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells. Nat Commun 8, 1456.
Information from this funding period will not be updated anymore. New research related information is available here.
Early detection and epigenetic rejuvenation strategies for age-associated cardiac dysfunctions in humans
Developmental and regenerative RNA biology

Institute for Neurophysiology & Center for Molecular Medicine Cologne
CMMC - PI - A 08
show more…
+49 221 478 89692
Institute for Neurophysiology & Center for Molecular Medicine Cologne
Robert-Koch-Str. 21
50931 Cologne
Deniz Bartsch (Post Doc)
Wenjie Yao (PhD student)
Kaustubh Kalamkar (PhD Student)
Arya Mesgary (MD-PhD)
Adrian Platen (Student intern)
Nicole Russ (Technician)
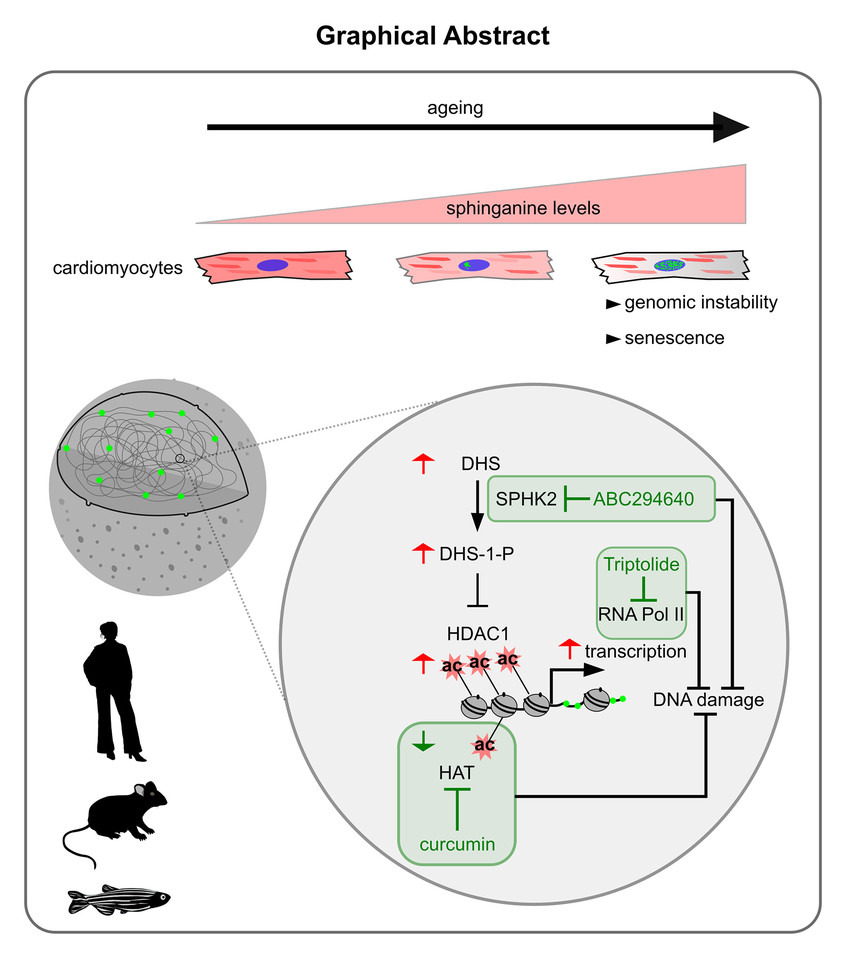
Fig. 2: Loss of genomic integrity induced by sphingolipid imbalance drives ageing in the heart:Graphical illustration summarizing the discovery of sphinganine (DHS) as an evolutionarily conserved hall-mark of cardiac ageing. Elevated DHS levels cause DNA damage in cardiomyocytes in vitro and in vivo. DHS derivative DHS1P is a potent HDAC inhibitor causing histone hyperacetylation and increased transcription, leading to DNA damage. Exogenous increase of DHS levels in young adult mice causes ageing‐associated functional impairment of the heart. Restoring histone acetylation levels by HAT inhibition prevents DHS‐induced cardiac DNA damage and functional impairment in vivo.