Despite the progress achieved in understanding the genetic basis of familial breast- and ovarian cancer, known genetic risk factors explain only less than half of all familial cases. Additional yet unknown genetic and non-genetic risk factors contribute to the multifactorial disease and modulate disease risk. Our goal is to identify novel risk factors and their interactions, followed by their implementation into personalised risk prediction and cancer prevention programmes.
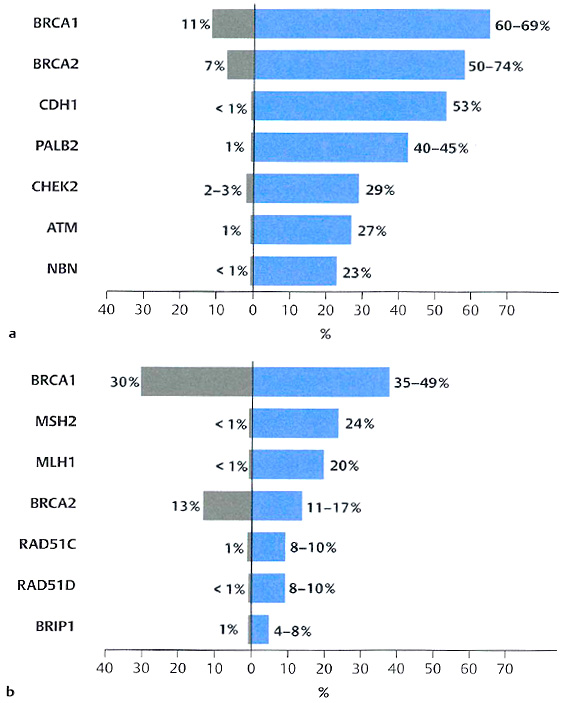
The Cologne Center for Familial Breast and Ovarian Cancer provides genetic counselling and support for women with an increased familial risk for breast cancer (BC) and ovarian cancer (OC), including interdisciplinary consultation and preventive measures. Despite the progress achieved in understanding the genetic basis of familial BC/OC, known genetic risk factors such as germline alterations in the known risk genes (Fig. 1) explain less than 50% of all familial cases (“missing heritability”). Thus, additional genetic and non-genetic risk factors contribute to the multifactorial disease and/or modulate disease risk. Our vision is to identify and validate novel risk factors, followed by their implementation into personalised risk prediction and cancer prevention programmes. To reach these goals we are closely cooperating with international study groups and coordinate the German Consortium for Hereditary Breast- and Ovarian Cancer (GC-HBOC), consisting of 17 national centers.
Within the framework of the GC-HBOC, more than 21.000 familial index patients were screened for pathogenic BRCA1/2 mutations with 24% being BRCA1/2-positive (Kast et al., 2016, J Med Genet). Besides BRCA1/2, additional known risk genes (Fig. 1) are only rarely mutated (<1%). Clinical data of these 21.000 patients (mutation status, family history, tumour histology, survival) is available for scientific use at the centralized national patient database operated by the IMISE (Institute for Medical Informatics, Statistics and Epidemiology, Leipzig). Today, our DNA biobank contains more than patient-derived 17.000 DNA samples readily available for research purposes, which is an important prerequisite for our studies.
We identified RAD51C as a novel OC risk gene which is, like BRCA1/2, involved in DNA repair. During this CMMC funding period, we identified FANCM as a novel risk gene for BC (Neidhardt et al., JAMA Oncology, accepted). Heterozygous loss of function mutations within the FANCM gene were significantly associated with familial BC risk, with an overall odds ratio (OR) of 2.05 and a mutation frequency of 1.03% in index cases. In familial BC patients with a BC onset before the age of 51 years, an elevated OR of 2.44 was observed. In a follow-up study, we show for the first time that the homozygous inactivation of FANCM results in a novel chromosomal instability syndrome (Catucci et al., in preparation). Functional analyses of FANCM missense alterations are ongoing. Moreover, we identified further candidates, with compelling evidence achieved for FANCC and GPRC5A as novel BC predisposition genes.
In cooperation with large international consortia BCAC (Breast Cancer Association Consortium) and CIMBA (Consortium of Investigators of Modifier Genes in BRCA1/2-Mutation Carriers) we participated in most genome-wide association studies (GWAS) that led to the identification of genetic variants which modify disease risk. The GC-HBOC is one of the largest contributing group with >8.500 DNA samples and pathology data provided. Up to date, we identified >200 low risk variants (SNPs) which modify disease risk, each of which is associated with a relative risk of 1.05 to 1.3 of developing BC. Our recent work suggests that a combined score based on these genotypes (“polygenic risk score”, PRS) could have substantial predictive value for risk stratification in the general population as well as in BRCA1/2 carriers. To determine whether such SNP panels could stratify women into clinically useful risk groups, we initiated a prospective validation of PRS findings in approximately 6.000 cases.
here is increasing evidence that BC/OC arising in BRCA1/2 germline mutation carriers are associated with a better response to DNA-damaging treatment regimen. This data prompted us to assess the germline mutation status of BC patients in the GeparSixto trial (NCT01426880). This trial assessed the efficacy of adding carboplatin to a regimen of paclitaxel, doxorubicin and bevacizumab for triple-negative BC patients. We show that the response rates are generally high in BRCA1/2 mutation carriers compared to non-carriers. Importantly, we demonstrate for the first time that a less intense treatment regimen should be considered for BRCA1/2 mutation carriers, which might be practice changing (Hahnen et al., JCO, under re-review).
Our current goals are i) to examine the utility of panels of SNPs (“polygenic risk score”) in the context of familial breast cancer, and ii) to further validate already identified novel BC risk genes (FANCM, FANCC, GPRC5A). However, there is ample evidence that novel risk genes are only rarely mutated, with expected mutation frequencies far below <1%. Consequently, the identification of these rarely mutated genes requires large cohorts. Thus, we joined two international projects (PERSPECTIVE, BRIDGES) focussing on large scale next generation sequencing approaches. Within the framewoork of the PERSPECTIVE project, the targeted sequencing of 130 novel candidate genes in 2.500 cases/2.5000 controls will we carried out in our laboratory in cooperation with the Cologne Center for Genomics (CCG).
Milne RL, …, Hahnen E, …, Schmutzler RK, …, Simard J (2016). Ten variants associated with risk of estrogen receptor negative breast cancer. Nat Genet. accepted
Dunning AM, …, Schmutzler RK, …, Edwards SL (2016). Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat Genet. Apr;48(4):374-86.
Rebbeck TR, …, Schmutzler RK, …, Hahnen E, …, Andrulis I. (2015). Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA;313(13):1347-61.
Kuchenbaecker KB, …, Schmutzler RK, …Chenevix-Trench G (2015): Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 47(2):164-71.
Day FR, …, Schmutzler RK, …, Murray A (2015). Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. Nov;47(11): 1294-303.
Michailidou K, …, Schmutzler RK, …, Easton DF (2015). Genome-wide association analysis ofmore than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. Apr;47(4):373-80.
Information from this funding period will not be updated anymore. New research related information is available here.

Center for Familial Breast- and Ovarian Cancer
CMMC - PI - assoc. RG 20
rita.schmutzler[at]uk-koeln.de
show more…+49 221 478 86509
+49 221 478 86510
Center for Familial Breast- and Ovarian Cancer
Kerpener Str. 34
50931 Cologne

Center for Familial Breast- and Ovarian Cancer
Co-Principal Investigator A 9 - Funding Period 2014-2016 since 2016 - member of Core Facuility Datenbank breast cancer
show more…+49 221 478 98409
+49 221 478 5949
Center for Familial Breast- and Ovarian Cancer
Kerpener Str. 34
50931 Cologne
http://familiaerer-brust-und-eierstockkrebs.uk-koeln.de/team/forschung-molekulare-gynaeko-onkologie
Jan Hauke (PostDoc)
Esther Pohl (PostDoc)
Julika Borde (doctoral student)
Guido Neidhardt (doctoral student)
Kristina Klaschik (doctoral student)
Nana Weber-Lassalle (doctoral student)
Konstantin Weber-Lassalle (doctoral student)
Sandra Kröber (technician)