The main focus of my lab is on dissecting the molecular basis of the structure-to-function relationship of mammalian genomes. We wish to understand how chromatin integrates different signaling stimuli to control transitions between homeostatic and deregulated gene expression programs via looping regulation – particularly how changes along the linear chromatin fibre translate into spatial networks. Deciphering the general rules that govern transcriptional homeostasis will allow us to predict how a cell might respond in disease, development, or ageing.
It is now well understood that mammalian genomes are much more than mere libraries where genetic information is stored. Due to paradigm-shifting technological advances over the last decade, we now also understand that genomes are four-dimensional entities, and should be studied in 3D space and over time in order to dissect their sequence-to-structure-to-function relationship. Our work aims at furthering our understanding of 3D genomic architecture and how this is affected by and aligned to transcriptional organization in nuclei.
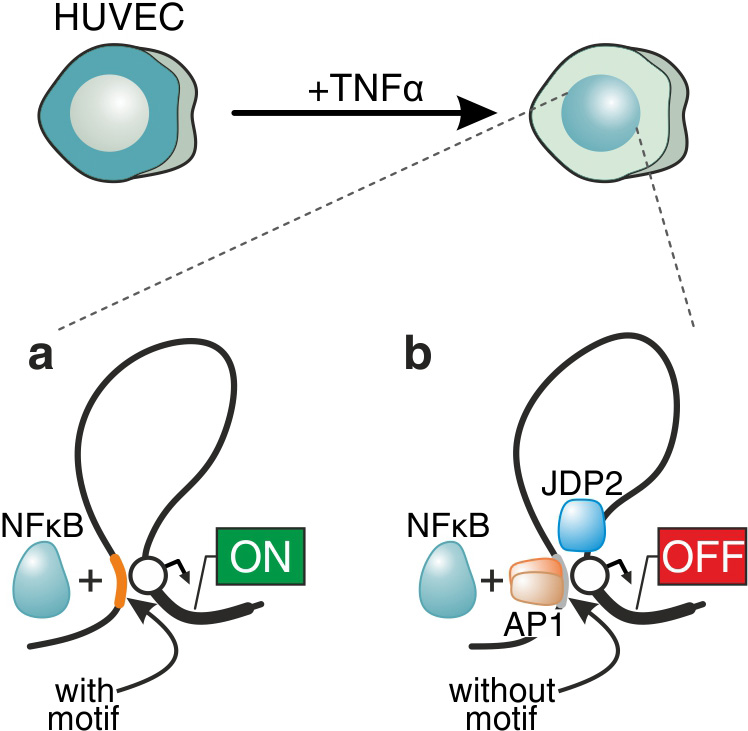
We perform comparative studies of inflammatory (TNF) versus migratory (TGF) signaling in primary endothelial cells, in order to understand how chromatin architecture allows for their diverse signaling dynamics (the former cascade unfolds within minutes, while the latter within hours). We focus on nucleosome and transcription factor occupancy landscapes, and on differences in transcriptional dynamics between the two cascades. We already described a global “priming” mechanism, whereby thousands of regions in the genome are rendered accessible following TNF stimulation, despite the absence of bound NF-κB (the factor driving the cascade); this mechanism also facilitates forthcoming changes in 3D chromatin conformation. We are currently in the process of finding out if “priming” is a universal means for preparing the genome for ensuing transcriptional programs. More recently, we discovered how non-canonical binding of transcription factors may alter their regulatory character – and turn an activator into a facilitator of gene repression (Figure 1). As a result, to further understand the molecular basis of these dynamics we initiated a project where we apply single-nucleotide resolution sequencing of nascent RNA (using our novel approach; Melnik et al, 2016), and complement it with single-cell RNA-seq (in collaboration with Karsten Rippe, Heidelberg) and single-molecule RNA FISH. As a whole, this work is bound to uncover fundamental differences in the transcriptional organization of mammalian nuclei in response to signaling and offer novel targets in these medically-relevant pathways.
We have been pursuing an integrative study of cellular senescence, the intrinsic mechanism that is responsible for the prevention of tumorigenesis, but also implicated in ageing. Here, we use different primary cell types (endothelial, lung fibroblast, and mesenchymal stem cells) in an effort to identify the common molecular backbone that signals entry into senescence. The molecular events, in 3D space and over time, driving the establishment of the senescent gene expression program remain largely unexplored, but are known to involve the activation of a proinflammatory transcriptional program. To this end, we drive the three cell types into replicative senescence, perform nascent RNA-seq, and seek to identify a common regulatory backbone between all three cell type-specific transcriptional programs, with an emphasis on differentially-regulated factors that associate with chromatin. We find genes coding for regulators of chromatin conformation being suppressed, and the levels of many transcriptional repressors being induced. Next, we use the whole-genome variant of the chromosome conformation capture technology, Hi-C (see Figure 2), to identify global changes in 3D chromatin organization. We have generated and analyzed >5 billion reads from proliferating and senescent cells, leading to interaction maps of <10-kbp resolution. Using these we identified marked reorganization at the multi-Mbp domain scale, a feature underpinned by the ubiquitous increase of the nuclear volume in senescent cells. The gain and loss of spatial interactions and the shift in topological domain boundaries correlates with non-histone chromatin constituents extruded from the nucleus into extracellular space, thus contributing to the senescence-associated secretory phenotype (SASP). Our work is the first to identify a link between the SASP and the reorganization of nuclear architecture, and it points to a particular nuclear checkpoint that prepares the cell’s response in the face of senescent arrest.
Our work on the connection between 3D chromatin folding and transcriptional organization focuses on either proinflammatory signaling, or on the onset and establishment of cellular senescence. These two processes share physiological and molecular commonalities, and their dissection will offer both a chance to identify novel effectors and potential therapeutic targets, and a deeper understanding of the 3D structure-to-function code of the human genome that still remains a challenge.
Michieletto D, Chiang M, Coli D, Papantonis A, Orlandini E, Cook PR, and Marenduzzo D (2018). Shaping epigenetic memory via genomic bookmarking. Nucleic Acids Res 46, 83-93.
Rada-Iglesias A, Grosveld FG, and Papantonis A (2018). Forces driving the three-dimensional folding of eukaryotic genomes. Mol Syst Biol 14, e8214.
Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmuller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nurnberg P, Marenduzzo D, Rippe K, and Papantonis A (2018). HMGB2 Loss upon Senescence Entry Disrupts Genomic Organization and Induces CTCF Clustering across Cell Types. Mol Cell 70, 730-744 e736.
Zirkel A, and Papantonis A (2018). Detecting Circular RNAs by RNA Fluorescence In Situ Hybridization. Methods Mol Biol 1724, 69-75.
Franzen J, Zirkel A, Blake J, Rath B, Benes V, Papantonis A, and Wagner W (2017). Senescence-associated DNA methylation is stochastically acquired in subpopulations of mesenchymal stem cells. Aging Cell 16, 183-191.
Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, Korhonen E, Smura T, Vapalahti O, Papantonis A, Schmidt-Chanasit J, Riparbelli M, Callaini G, Kronke M, Utermohlen O, and Gopalakrishnan J (2017). Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell 20, 397-406 e395.
Nikolic M, Papantonis A, and Rada-Iglesias A (2017). GARLIC: a bioinformatic toolkit for aetiologically connecting diseases and cell type-specific regulatory maps. Hum Mol Genet 26, 742-752.
Brant L, Georgomanolis T, Nikolic M, Brackley CA, Kolovos P, van Ijcken W, Grosveld FG, Marenduzzo D, and Papantonis A (2016). Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Mol Syst Biol 12, 891.
Georgomanolis T, Sofiadis K, and Papantonis A (2016). Cutting a Long Intron Short: Recursive Splicing and Its Implications. Front Physiol 7, 598.
Kolovos P, Georgomanolis T, Koeferle A, Larkin JD, Brant L, Nikolic M, Gusmao EG, Zirkel A, Knoch TA, Costa IG, van Ijcken WF, Cook PR, Grosveld FG, and Papantonis A (2016). Binding of nuclear factor kappa B to non-canonical consensus sites reveals its multimodal role during the early inflammatory response. Genome Res 26, 1478-1489.
Melnik S, Caudron-Herger M, Brant L, Carr IM, Rippe K, Cook PR, and Papantonis A (2016). Isolation of the protein and RNA content of active sites of transcription from mammalian cells. Nat Protoc 11, 553-565.
Brant L, and Papantonis A (2015). Contribution of 3D chromatin architecture to the maintenance of pluripotency. Curr Stem Cell Rep 1, 1-6.
Caudron-Herger M, Cook PR, Rippe K, and Papantonis A (2015). Dissecting the nascent human transcriptome by analysing the RNA content of transcription factories. Nucleic Acids Res 43, e95.
Kelly S, Georgomanolis T, Zirkel A, Diermeier S, O'Reilly D, Murphy S, Langst G, Cook PR, and Papantonis A (2015). Splicing of many human genes involves sites embedded within introns. Nucleic Acids Res 43, 4721-4732.
Kelly S, Greenman C, Cook PR, and Papantonis A (2015). Exon Skipping Is Correlated with Exon Circularization. J Mol Biol 427, 2414-2417.
Papantonis A, Swevers L, and Iatrou K (2015). Chorion genes: a landscape of their evolution, structure, and regulation. Annu Rev Entomol 60, 177-194.
Diermeier S, Kolovos P, Heizinger L, Schwartz U, Georgomanolis T, Zirkel A, Wedemann G, Grosveld F, Knoch TA, Merkl R, Cook PR, Langst G, and Papantonis A (2014). TNFalpha signalling primes chromatin for NF-kappaB binding and induces rapid and widespread nucleosome repositioning. Genome Biol 15, 536.
Zirkel A, and Papantonis A (2014). Transcription as a force partitioning the eukaryotic genome. Biol Chem 395, 1301-1305.
Brackley CA, Taylor S, Papantonis A, Cook PR, and Marenduzzo D (2013). Nonspecific bridging-induced attraction drives clustering of DNA-binding proteins and genome organization. Proc Natl Acad Sci U S A 110, E3605-3611.
Larkin JD, Papantonis A, and Cook PR (2013). Promoter type influences transcriptional topography by targeting genes to distinct nucleoplasmic sites. J Cell Sci 126, 2052-2059.
Larkin JD, Papantonis A, Cook PR, and Marenduzzo D (2013). Space exploration by the promoter of a long human gene during one transcription cycle. Nucleic Acids Res 41, 2216-2227.
Papantonis A, and Cook PR (2013). Transcription factories: genome organization and gene regulation. Chem Rev 113, 8683-8705.
Ahuja, G., Bartsch, D., Yao, W., Geissen, S., Frank, S., Aguirre, A., Russ, N., Messling, J.E., Dodzian, J., Lagerborg, K.A., Vargas, N.E., Muck, J.S., Brodesser, S., Baldus, S., Sachinidis, A., Hescheler, J., Dieterich, C., Trifunovic, A., Papantonis, A., Petrascheck, M., Klinke, A., Jain, M., Valenzano, D.R., and Kurian, L. (2019). Loss of genomic integrity induced by lysosphingolipid imbalance drives ageing in the heart. EMBO Rep 20.
Frank, S., Ahuja, G., Bartsch, D., Russ, N., Yao, W., Kuo, J.C., Derks, J.P., Akhade, V.S., Kargapolova, Y., Georgomanolis, T., Messling, J.E., Gramm, M., Brant, L., Rehimi, R., Vargas, N.E., Kuroczik, A., Yang, T.P., Sahito, R.G.A., Franzen, J., Hescheler, J., Sachinidis, A., Peifer, M., Rada-Iglesias, A., Kanduri, M., Costa, I.G., Kanduri, C., Papantonis, A., and Kurian, L. (2019). yylncT Defines a Class of Divergently Transcribed lncRNAs and Safeguards the T-mediated Mesodermal Commitment of Human PSCs. Cell Stem Cell 24, 318-27 e8.
Gothe, H.J., Bouwman, B.A.M., Gusmao, E.G., Piccinno, R., Petrosino, G., Sayols, S., Drechsel, O., Minneker, V., Josipovic, N., Mizi, A., Nielsen, C.F., Wagner, E.M., Takeda, S., Sasanuma, H., Hudson, D.F., Kindler, T., Baranello, L., Papantonis, A., Crosetto, N., and Roukos, V. (2019). Spatial Chromosome Folding and Active Transcription Drive DNA Fragility and Formation of Oncogenic MLL Translocations. Mol Cell 75, 267-83 e12.
Lalioti, M.E., Arbi, M., Loukas, I., Kaplani, K., Kalogeropoulou, A., Lokka, G., Kyrousi, C., Mizi, A., Georgomanolis, T., Josipovic, N., Gkikas, D., Benes, V., Politis, P.K., Papantonis, A., Lygerou, Z., and Taraviras, S. (2019). GemC1 governs multiciliogenesis through direct interaction with and transcriptional regulation of p73. J Cell Sci 132.
Lalioti, M.E., Kaplani, K., Lokka, G., Georgomanolis, T., Kyrousi, C., Dong, W., Dunbar, A., Parlapani, E., Damianidou, E., Spassky, N., Kahle, K.T., Papantonis, A., Lygerou, Z., and Taraviras, S. (2019). GemC1 is a critical switch for neural stem cell generation in the postnatal brain. Glia10.1002/glia.23690.
Michieletto D, Chiang M, Coli D, Papantonis A, Orlandini E, Cook PR, and Marenduzzo D (2018). Shaping epigenetic memory via genomic bookmarking. Nucleic Acids Res 46, 83-93.
Rada-Iglesias A, Grosveld FG, and Papantonis A (2018). Forces driving the three-dimensional folding of eukaryotic genomes. Mol Syst Biol 14, e8214.
Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmuller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nurnberg P, Marenduzzo D, Rippe K, and Papantonis A (2018). HMGB2 Loss upon Senescence Entry Disrupts Genomic Organization and Induces CTCF Clustering across Cell Types. Mol Cell 70, 730-744 e736.
Zirkel A, and Papantonis A (2018). Detecting Circular RNAs by RNA Fluorescence In Situ Hybridization. Methods Mol Biol 1724, 69-75.
Franzen J, Zirkel A, Blake J, Rath B, Benes V, Papantonis A, and Wagner W (2017). Senescence-associated DNA methylation is stochastically acquired in subpopulations of mesenchymal stem cells. Aging Cell 16, 183-91.
Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, Korhonen E, Smura T, Vapalahti O, Papantonis A, Schmidt-Chanasit J, Riparbelli M, Callaini G, Kronke M, Utermohlen O, and Gopalakrishnan J (2017). Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell 20, 397-406 e5.
Nikolic M, Papantonis A, and Rada-Iglesias A (2017). GARLIC: a bioinformatic toolkit for aetiologically connecting diseases and cell type-specific regulatory maps. Hum Mol Genet 26, 742-52.
Information from this funding period will not be updated anymore. New research related information is available here.
Towards a structure-to-function code for mammalian genomes

Institute of Pathology - University Medical Center Göttingen
CMMC - assoc. RG 26 (former CMMC JRG Leader)
argyris.papantonis[at]med.uni-goettingen.de
show more…+49 551 39 65734
Institute of Pathology - University Medical Center Göttingen
present adress: University Medical Center Göttingen Robert-Koch-Strasse 40
37075 Göttingen
http://zmmk-sbc.uni-koeln.de/JRG_VIII___Systems_Biology_of_Chromatin/People.html
CMMC Profile Page
Anne Zirkel (PostDoc)
Athanasia Mizi (PostDoc)
Yulia Kargapolova (PostDoc)
Lilija Brant (doctoral student)
Milos Nikolic (doctoral student)
Konstantinos Sofiadis (doctoral student)
Theodore Georgomanolis (technician)
Ruiyuan Zheng (PhD student)